Abstract
Title: Effects on Ubiquitination Pathway in Methionine Deprived Prostate Cancer Cell Lines
Authors: Robert Iandoli, OMS II, Andy Aleman, OMS II, Trevor Fuhriman, OMS II, KV Venkatachalam, PhD
Program: Dr. Kiran C. Patel College of Osteopathic Medicine, Nova Southeastern University
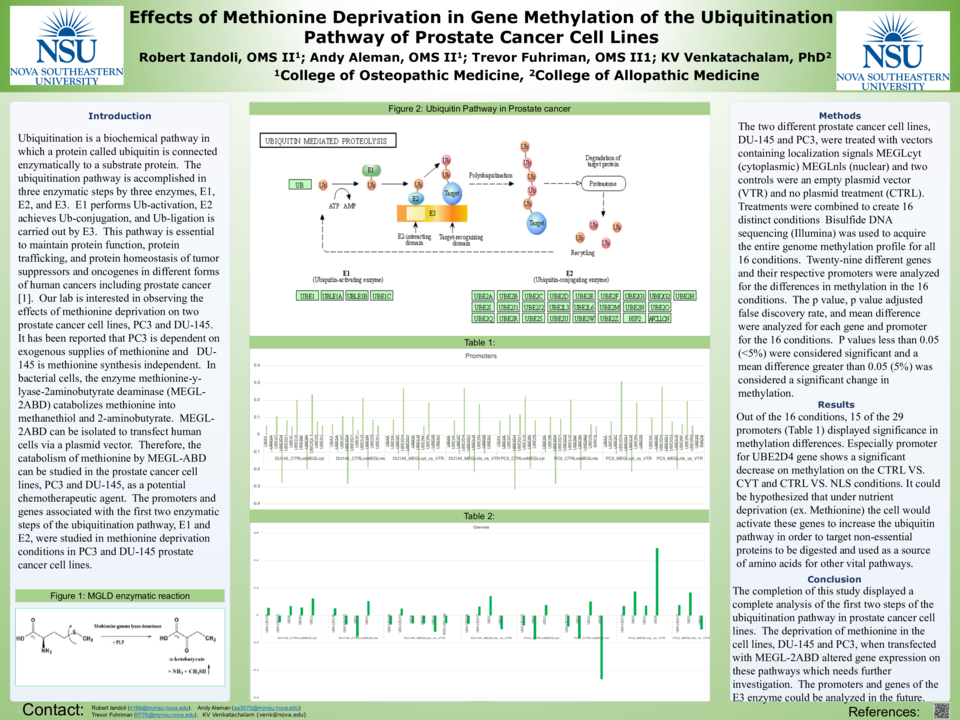
Background: Ubiquitination is a biochemical pathway in which a protein called ubiquitin is connected enzymatically to a substrate protein. The ubiquitination pathway is accomplished in three enzymatic steps by three enzymes, E1, E2, and E3. E1 performs Ub-activation, E2 achieves Ub-conjugation, and Ub-ligation is carried out by E3. This pathway is essential to maintain protein function, protein trafficking, and protein homeostasis of tumor suppressors and oncogenes in different forms of human cancers including prostate cancer [1]. Our lab is interested in observing the effects of methionine deprivation on two prostate cancer cell lines, PC3 and DU-145. It has been reported that PC3 is dependent on exogenous supplies of methionine and DU-145 is methionine synthesis independent. In bacterial cells, the enzyme methionine-y-lyase-2aminobutyrate deaminase (MEGL-2ABD) catabolizes methionine into methanethiol and 2-aminobutyrate. MEGL-2ABD can be isolated to transfect human cells via a plasmid vector. Therefore, the catabolism of methionine by MEGL-ABD can be studied in the prostate cancer cell lines, PC3 and DU-145, as a potential chemotherapeutic agent. The promoters and genes associated with the first two enzymatic steps of the ubiquitination pathway, E1 and E2, were studied in methionine deprivation conditions in PC3 and DU-145 prostate cancer cell lines.
Objective: The objective of these studies is to determine if there are potential chemotherapeutic targets located in the first two steps, E1 and E2, of the ubiquitination pathway of the two prostate cancer cell lines, PC3 and DU-145.
Methods: The two different prostate cancer cell lines, DU-145 and PC3, were treated with vectors containing localization signals MEGLcyt (cytoplasmic) MEGLnls (nuclear) and two controls were an empty plasmid vector (VTR) and no plasmid treatment (CTRL). Treatments were combined to create 16 distinct conditions Bisulfide DNA sequencing (Illumina) was used to acquire the entire genome methylation profile for all 16 conditions. Twenty-nine different genes and their respective promoters were analyzed for the differences in methylation in the 16 conditions. The p value, p value adjusted false discovery rate, and mean difference were analyzed for each gene and promoter for the 16 conditions. P values less than 0.05 (<5%) were considered significant and a mean difference greater than 0.05 (5%) was considered a significant change in methylation.
Results: Out of the 16 conditions, 15 of the 29 promoters (UBA3, UBE2A, UBE2C, UBE2D4, UBE2G1, UBE2G2, UBE2I, UBE2J2, UBE2M UBE2N, UBE2S, UBE2U, UBE2M, UBE2QL1, UBE2R2, BIRC6) and 8 of the 29 genes (UBA2, UBA3, UBE2B, UBE2E3, UBE2J1, UBE2C, UBE2N, BIRC6) displayed significance in methylation differences.
Conclusion: The completion of this study displayed a complete analysis of the first two steps of the ubiquitination pathway in prostate cancer cell lines. The deprivation of methionine in the cell lines, DU-145 and PC3, when transfected with MEGL-2ABD altered gene expression.